OpenOnDemand
OpenOnDemand is a platform for running computational tasks on a cluster from a web browser. If those tasks are interactive, it provides the ability to interact with them once the task has started its execution. OpenOnDemand has an "App" based plugin system for adding new types of computational tasks and interactivity.
One of the greatest benefits of this system is remote access to large machines for computational tasks, without the need for utilizing a commnand-line interface that is difficult to learn.
Our installation is available at https://ondemand.beocat.ksu.edu
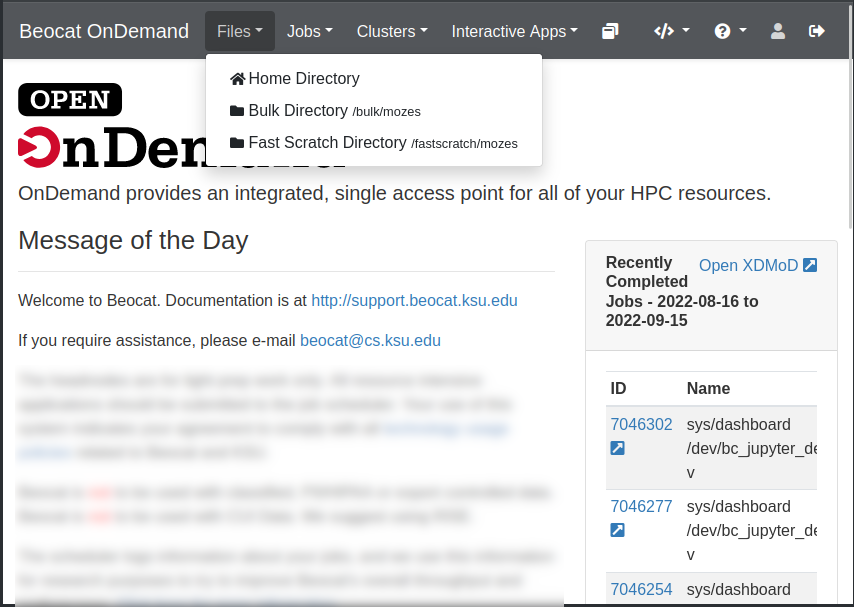
File Management
File management can be accessed through the Files dropdown in the dashboard.
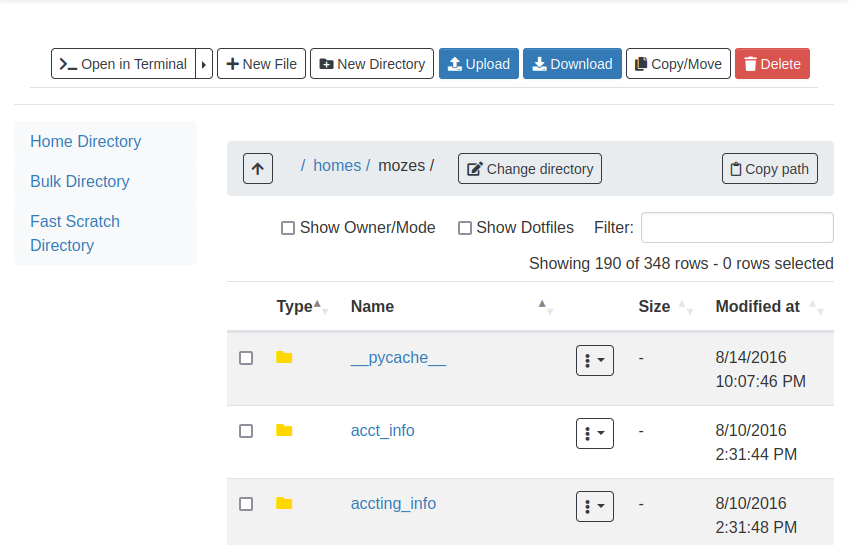
Once you click on Home Directory, you can manage your files, upload/download/rename/edit and view.
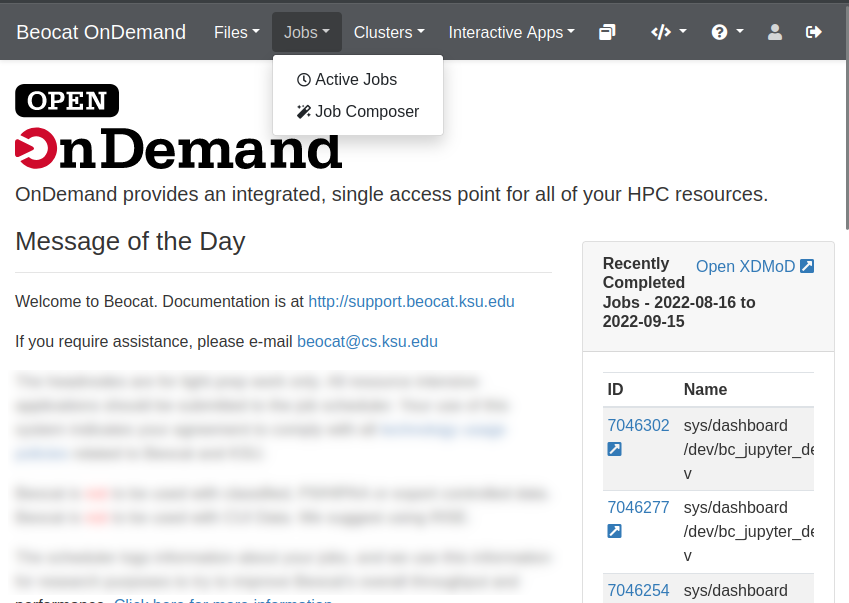
Job Management
A cluster isn't much of a cluster if it can't run jobs for you to lookup later. OpenOnDemand has a robust job management application builtin.
It is accessible from the Jobs dropdown in the dashboard.
View Active Jobs
You can view your active jobs and get more information about them from the Active Jobs option in the Jobs dropdown
Active jobs app in OpenOnDemand
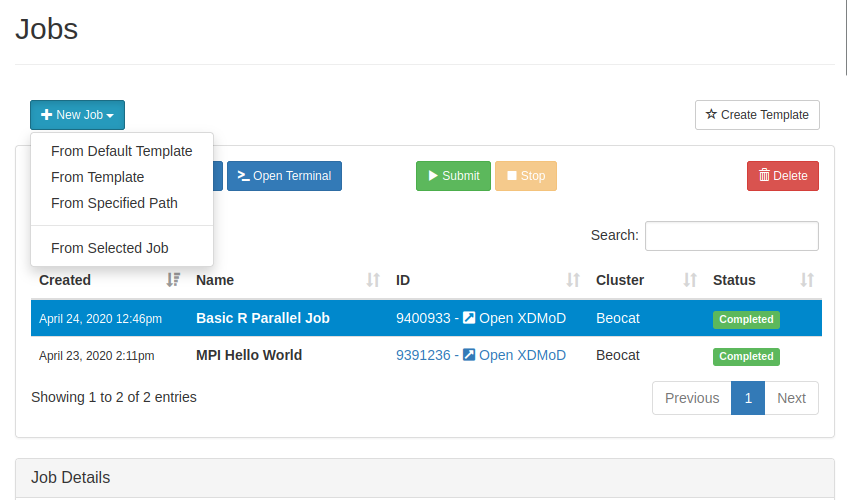
Compose Jobs
You can create new jobs through the "Job Composer" in the jobs dropdown.
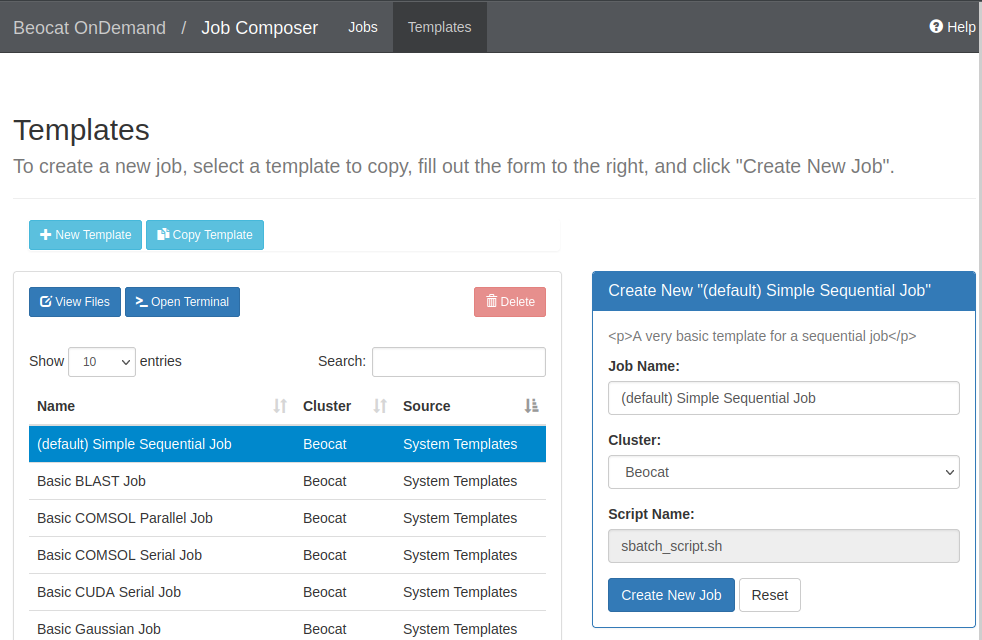
If we create a new job from a template, you're given a list of templates to use:
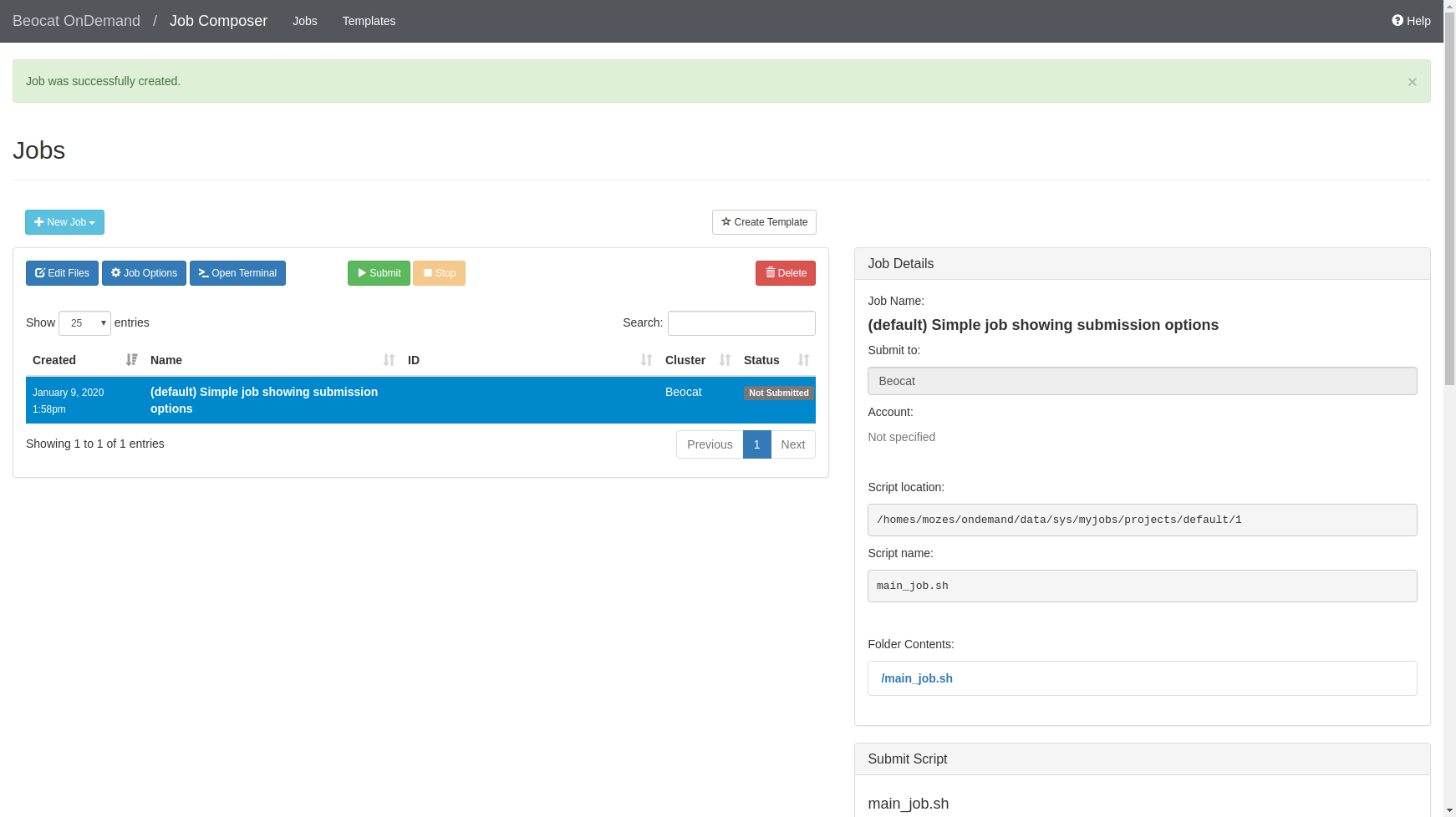
If we choose the default template, you can run it, or edit the job script to make it do what you would like
RStudio
RStudio is one of the interactive applications that we've enabled for use within OpenOnDemand
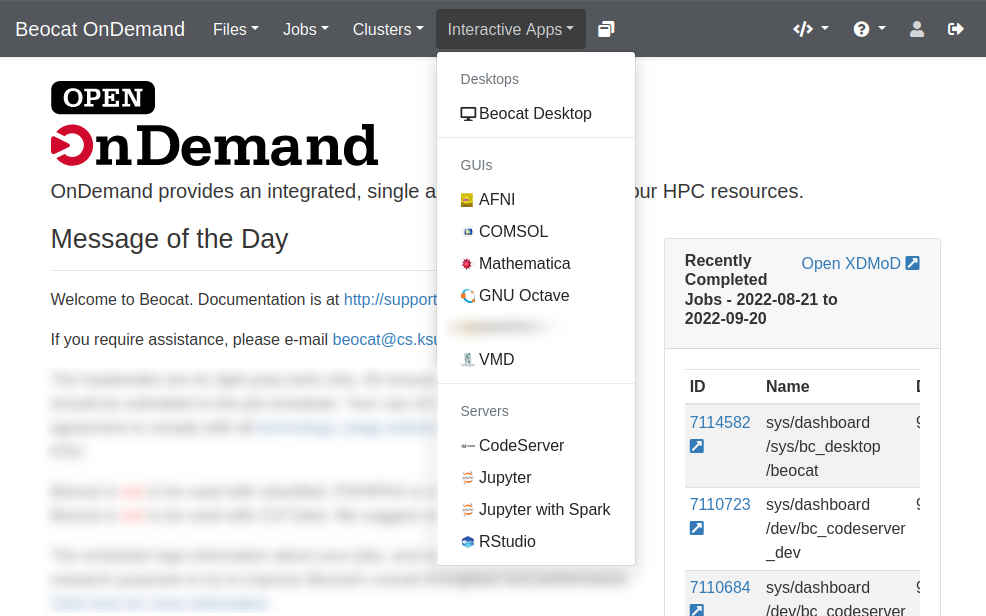
You launch interactive apps through the "Interactive Apps" dropdown.
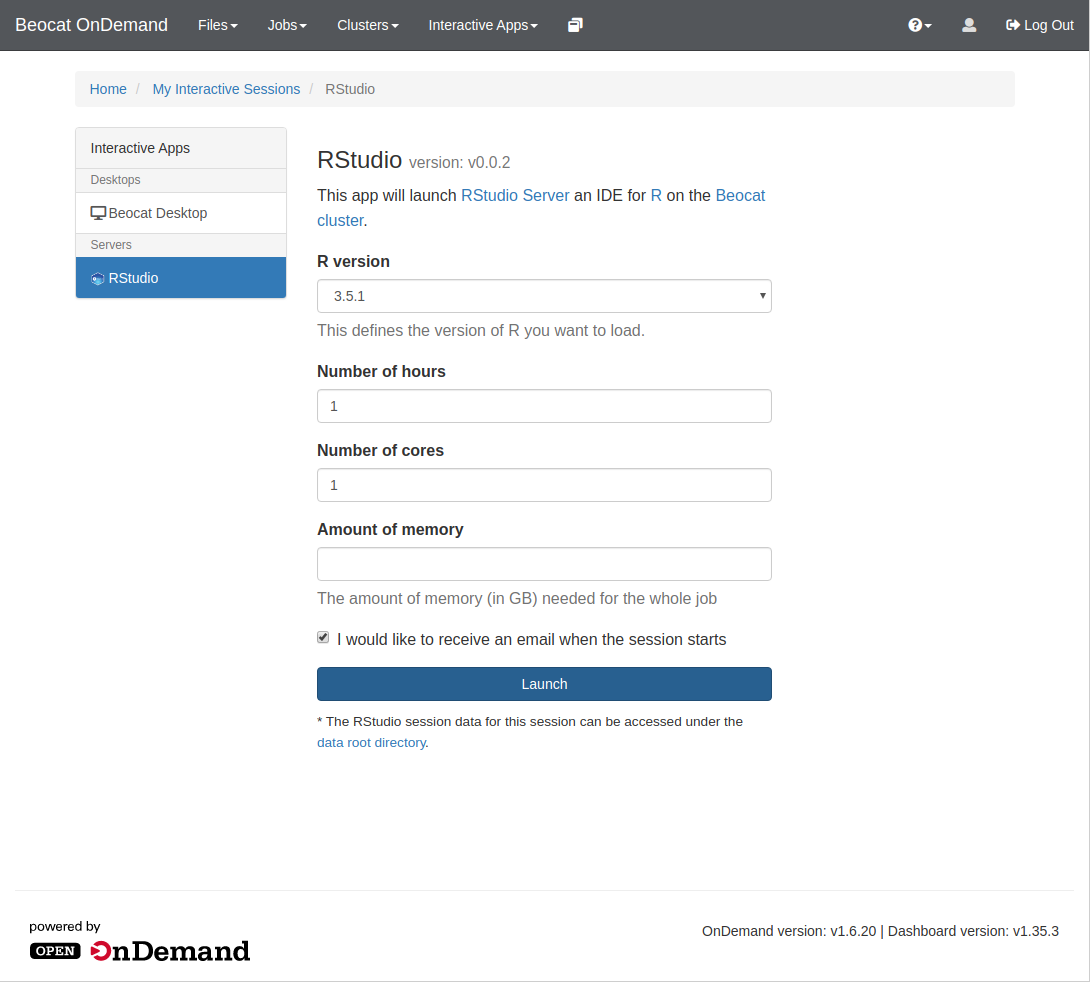
Once you click on RStudio, you'll be brought to a page allowing you to specify requirements for your RStudio run, e.g. memory, cores, and runtime.
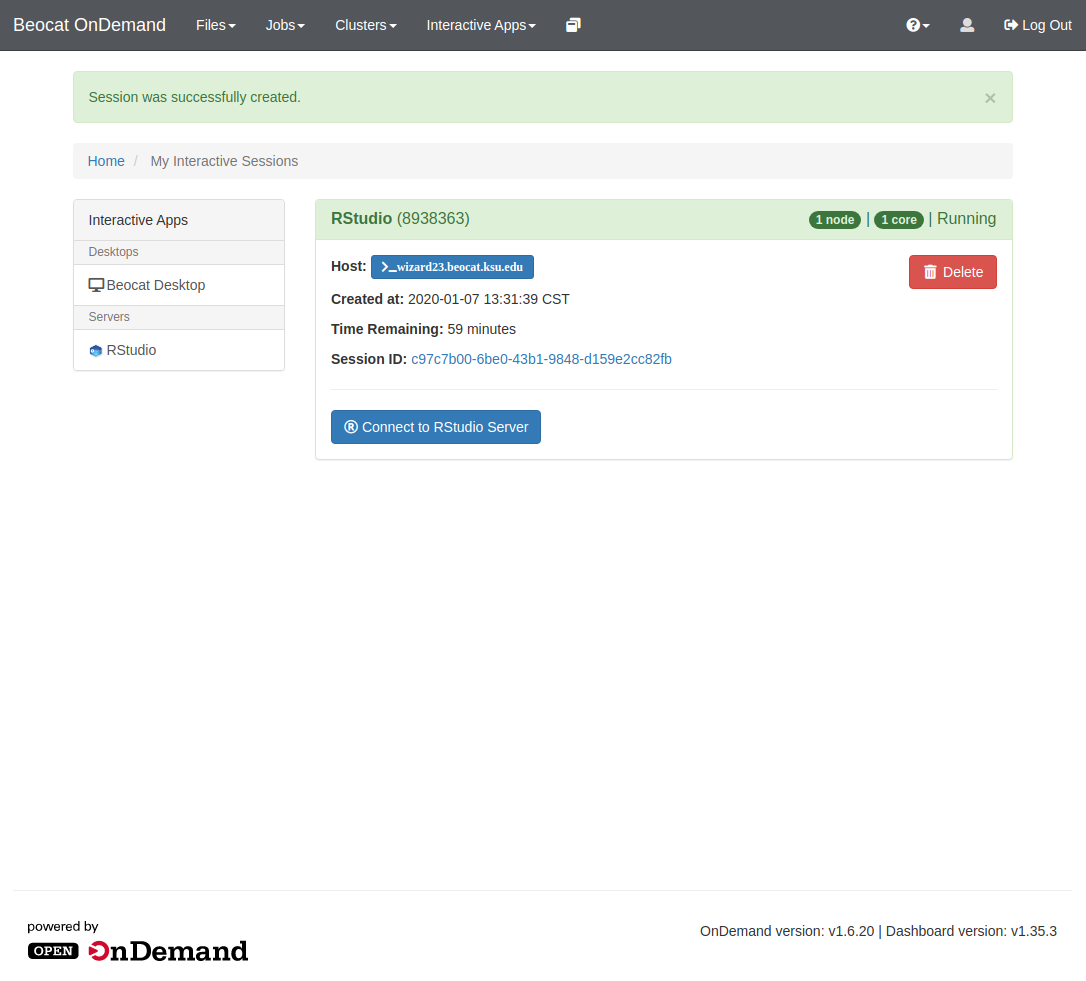
Once the job is submitted, the scheduler will take it and run it when space is available. Once the job is running, "My Interactive sessions" page will look like this:
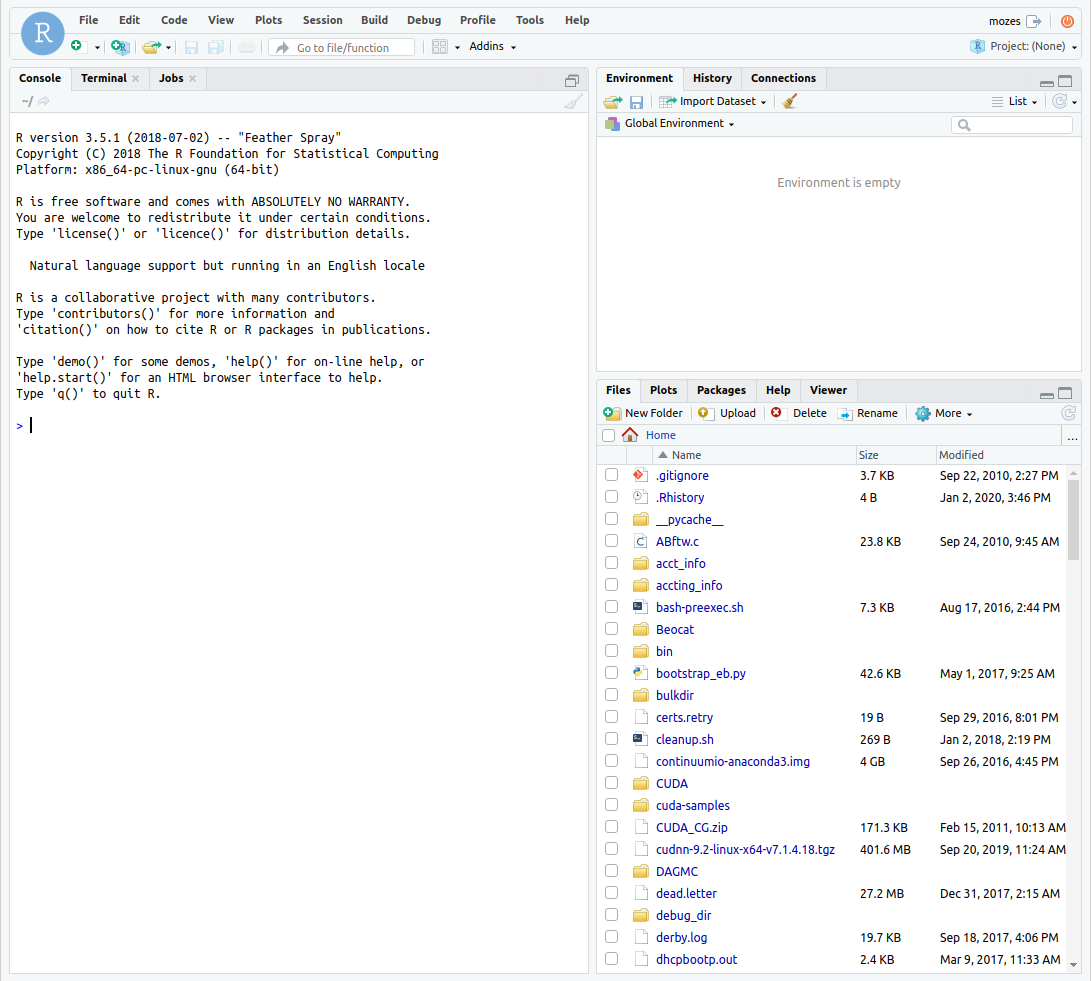
From there, you can connect to RStudio and it will bring you to a familiar interface.
Jupyter
Beocat Desktop
Shell Access
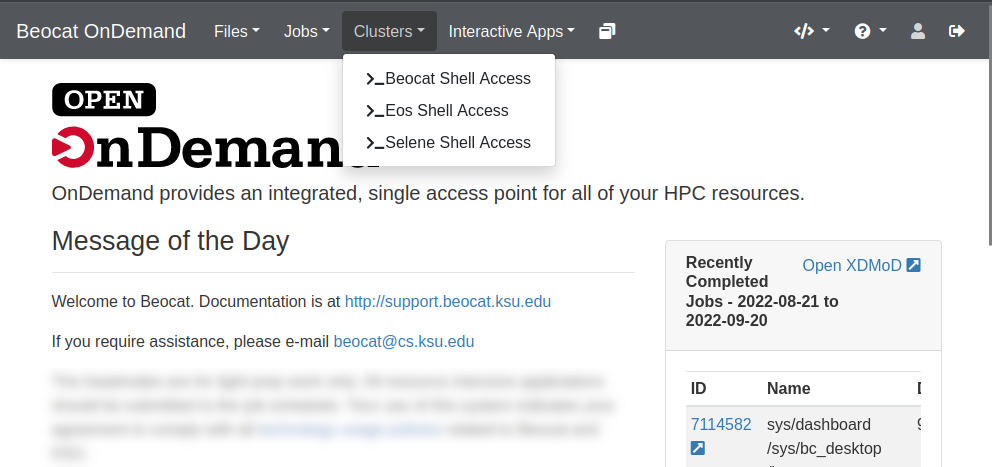
Somethings, no matter how hard we try, are easier to do via the command line. OpenOnDemand also gives you a way to handle those cases via the Clusters dropdown
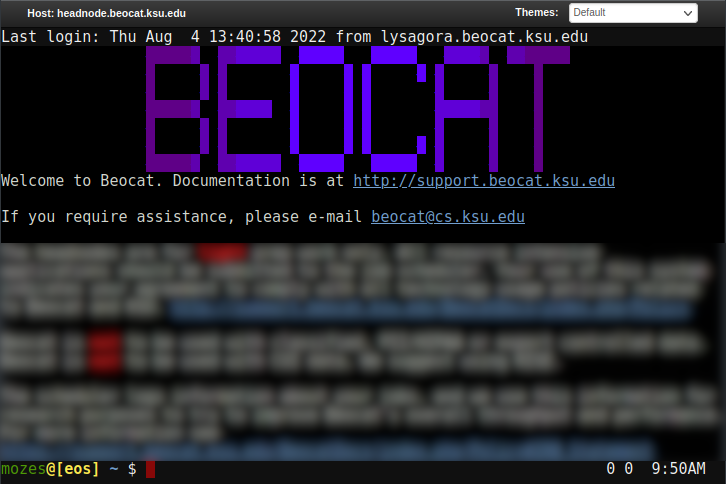
You can choose an individual headnode, if need-be, or you can choose "Beocat Shell Access" to be given one of the headnodes at random. Once chosen, you should be able to have a familiar command-line experience