Nathanrwells (talk | contribs) |
Nathanrwells (talk | contribs) |
||
| (20 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
== How do I access Galaxy? == | == How do I access Galaxy? == | ||
Access to Beocat's local instance of Galaxy is easy. Simply navigate to [https://galaxy.beocat.ksu.edu/ https://galaxy.beocat.ksu.edu/] and sign in if prompted using the Keycloak login. This will use your Beocat EID and Password to login, you will also be prompted to authenticate against DUO. | Access to Beocat's local instance of Galaxy is easy. Simply navigate to [https://galaxy.beocat.ksu.edu/ https://galaxy.beocat.ksu.edu/] and sign in if prompted using the Keycloak login. This will use your Beocat EID and Password to login, you will also be prompted to authenticate against DUO. | ||
Please note that this utilizes Beocat's /bulk directory. This is a billed directory, and as such, if usage in your respective upload directory for Galaxy exceeds the billing threshold (1T), we will contact you regarding remediation. | |||
== Upload Larger Files to Galaxy == | |||
Have some larger files that need to be uploaded to Galaxy? We provide some documentation on how to upload files directly to Beocat and then import them to Galaxy for use. It can be found here: [[Galaxy_File_Upload| Galaxy File Upload]] | |||
== How do I use Galaxy? == | |||
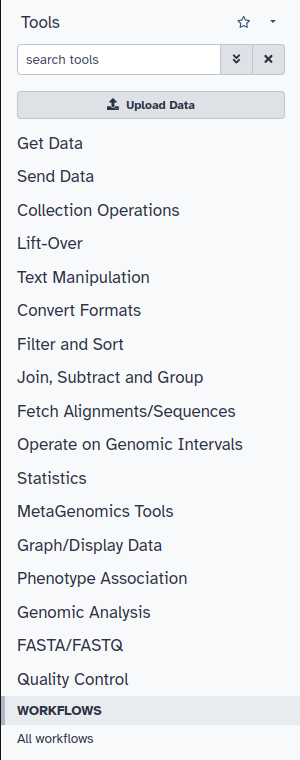
After accessing our local instance of Galaxy you should have access to all installed tools which will be in the tool panel on the left-hand side of your home screen on Galaxy. It should look like this: | |||
[[File:Galaxy Toolbox.png|Tool Panel]] | |||
Each category (Get Data, Send Data, MetaGenomics Tools, etc) is expandable reveal subcategories that help sort out many installed tools. Each tool will be placed under its respective category once it is installed. | |||
From here you can select a tool to use and submit to the Slurm cluster. After opening a tool you will be given options (if available) to configure the tool, including its inputs, what the tool will do (within its constraints) outputs, and what compute resources the tool will submit with. This means that you can specify the resources given to slurm jobs by expanding the drop-down menu "Job Resource Paramters" and selecting "Specify Job resource parameters" which will show the following options to modify. | |||
<B>Processors</B>: Number of processing cores, 'ppn' value (1-128) this is equivalent to slurms "--cpus-per-task".<br> | |||
<B>Memory</B>: Memory size in gigabytes, 'pmem' value (1-1500). Note that the job scheduler uses --mem-per-cpu to allocate memory for your slurm job. This mean the number given for memory will be multiplied by the Processors count from above. I.e 2 Processors with 5 Memory will be 10GB of memory.<br> | |||
<B>Priority Queue</B>: If you have access to a priority queue, and would like to use it, enter the partition name here.<br> | |||
<B>Runtime</B>: How long you want your job to run for in hours. | |||
If you fail to switch your job resource parameters, your job will submit with the default resources. This means it will submit with 5Gb of Memory and 1 CPU with 1 hour of Runtime and no Priority queue. | |||
=== Canceling a running job === | |||
While Galaxy does submit to slurm, you will not be able to cancel the job in the same way you typically do. With Galaxy, to cancel your upcoming/currently running job simply press the trash can icon next the name of your job. | |||
== Tool Requests == | == Tool Requests == | ||
If you are missing a specific tool and would like to have it added to Galaxy, please contact | If you are missing a specific tool and would like to have it added to Galaxy, please contact Beocat staff through a [https://support.ksu.edu/TDClient/30/Portal/Requests/ServiceDet?ID=44 TDX Ticket] with a link to the tool. Additionally, you can browse through Galaxy's own [https://toolshed.g2.bx.psu.edu/ toolshed] to make a recommendation. | ||
== Data Management == | |||
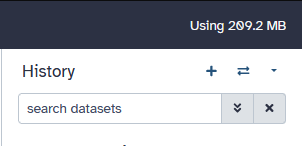
Galaxy follows the typical costs for bulk data storage, as Galaxy utilizes /bulk/galaxy for storage. Bulk data storage may be provided at a cost of $45/TB/year billed monthly. Billing starting at 1TB of usage. Users can easily see how much data they are utilizing in galaxy by checking the top right corner of the 'home' page of galaxy. This will say "Using ####MB/GB/TB", above your histories. | |||
[[File:Galaxy_Data_usage_example.png|Usage Data]] | |||
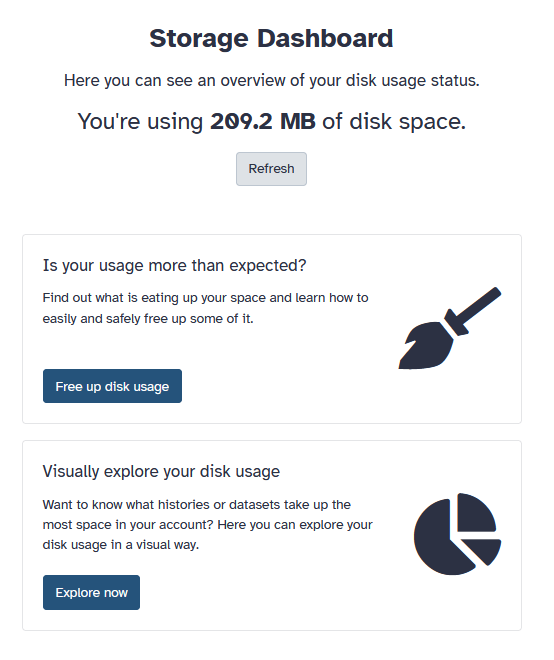
Clicking on this usage will bring you to a storage dashboard where you can easily manage your files and derelict dataset histories. | |||
[[File:Storage_dashboard.png|Storage Dashboard]] | |||
== Requesting Help == | == Requesting Help == | ||
To request help with [https://galaxy.beocat.ksu.edu/ https://galaxy.beocat.ksu.edu/], please contact | To request help with [https://galaxy.beocat.ksu.edu/ https://galaxy.beocat.ksu.edu/], please contact Beocat staff through a [https://support.ksu.edu/TDClient/30/Portal/Requests/ServiceDet?ID=44 TDX Ticket].<br> | ||
When requesting help, it is best to give as much information as possible so that we may solve your issue in a timely manner. | When requesting help, it is best to give as much information as possible so that we may solve your issue in a timely manner. | ||
Latest revision as of 07:46, 26 June 2025
What is Galaxy?
Galaxy is a scientific workflow, data integration, and data and analysis persistence and publishing platform that aims to make computational biology accessible to research scientists that do not have computer programming or systems administration experience.
How do I access Galaxy?
Access to Beocat's local instance of Galaxy is easy. Simply navigate to https://galaxy.beocat.ksu.edu/ and sign in if prompted using the Keycloak login. This will use your Beocat EID and Password to login, you will also be prompted to authenticate against DUO.
Please note that this utilizes Beocat's /bulk directory. This is a billed directory, and as such, if usage in your respective upload directory for Galaxy exceeds the billing threshold (1T), we will contact you regarding remediation.
Upload Larger Files to Galaxy
Have some larger files that need to be uploaded to Galaxy? We provide some documentation on how to upload files directly to Beocat and then import them to Galaxy for use. It can be found here: Galaxy File Upload
How do I use Galaxy?
After accessing our local instance of Galaxy you should have access to all installed tools which will be in the tool panel on the left-hand side of your home screen on Galaxy. It should look like this:
Each category (Get Data, Send Data, MetaGenomics Tools, etc) is expandable reveal subcategories that help sort out many installed tools. Each tool will be placed under its respective category once it is installed.
From here you can select a tool to use and submit to the Slurm cluster. After opening a tool you will be given options (if available) to configure the tool, including its inputs, what the tool will do (within its constraints) outputs, and what compute resources the tool will submit with. This means that you can specify the resources given to slurm jobs by expanding the drop-down menu "Job Resource Paramters" and selecting "Specify Job resource parameters" which will show the following options to modify.
Processors: Number of processing cores, 'ppn' value (1-128) this is equivalent to slurms "--cpus-per-task".
Memory: Memory size in gigabytes, 'pmem' value (1-1500). Note that the job scheduler uses --mem-per-cpu to allocate memory for your slurm job. This mean the number given for memory will be multiplied by the Processors count from above. I.e 2 Processors with 5 Memory will be 10GB of memory.
Priority Queue: If you have access to a priority queue, and would like to use it, enter the partition name here.
Runtime: How long you want your job to run for in hours.
If you fail to switch your job resource parameters, your job will submit with the default resources. This means it will submit with 5Gb of Memory and 1 CPU with 1 hour of Runtime and no Priority queue.
Canceling a running job
While Galaxy does submit to slurm, you will not be able to cancel the job in the same way you typically do. With Galaxy, to cancel your upcoming/currently running job simply press the trash can icon next the name of your job.
Tool Requests
If you are missing a specific tool and would like to have it added to Galaxy, please contact Beocat staff through a TDX Ticket with a link to the tool. Additionally, you can browse through Galaxy's own toolshed to make a recommendation.
Data Management
Galaxy follows the typical costs for bulk data storage, as Galaxy utilizes /bulk/galaxy for storage. Bulk data storage may be provided at a cost of $45/TB/year billed monthly. Billing starting at 1TB of usage. Users can easily see how much data they are utilizing in galaxy by checking the top right corner of the 'home' page of galaxy. This will say "Using ####MB/GB/TB", above your histories.
Clicking on this usage will bring you to a storage dashboard where you can easily manage your files and derelict dataset histories.
Requesting Help
To request help with https://galaxy.beocat.ksu.edu/, please contact Beocat staff through a TDX Ticket.
When requesting help, it is best to give as much information as possible so that we may solve your issue in a timely manner.
Acknowledgements
Beocat's installation of UseGalaxy is funded through K-INBRE with an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103418.
This initiative was started through the Data Science Core group to bring easy to use GUI-based computational biology research to students and researchers at Kansas State University through Beocat.
Additional information on K-INBRE can be found here