No edit summary |
Nathanrwells (talk | contribs) |
||
| (24 intermediate revisions by 3 users not shown) | |||
| Line 5: | Line 5: | ||
interactivity. | interactivity. | ||
One of the greatest benefits of this system is remote access to large machines for computational tasks, without the need for utilizing a | One of the greatest benefits of this system is remote access to large machines for computational tasks, without the need for utilizing a command-line interface that is difficult to learn. | ||
Our installation is available at https://ondemand.beocat.ksu.edu | Our installation is available at https://ondemand.beocat.ksu.edu | ||
| Line 17: | Line 17: | ||
[[File:Ood Files Launch.png|The OpenOnDemand File management application]] | [[File:Ood Files Launch.png|The OpenOnDemand File management application]] | ||
If you're looking for a way to get your files into and out of OneDrive, Google Drive, other cloud providers you may be interested in taking a look at our documentation for [[Onedrive Data Transfer]] | |||
=== Job Management === | === Job Management === | ||
A cluster isn't much of a cluster if it can't run jobs for you to lookup later. OpenOnDemand has a robust job management application builtin. | |||
It is accessible from the Jobs dropdown in the dashboard. | |||
[[File:OOD JOBS DROPDOWN ACTIVE.png|Screenshot of the Jobs dropdown in the openondemand dashboard]] | |||
==== View Active Jobs ==== | |||
You can view your active jobs and get more information about them from the Active Jobs option in the Jobs dropdown | |||
[[File:OOD JOBS ACTIVE.png|Active jobs app in OpenOnDemand]] | |||
==== Compose Jobs ==== | |||
You can create new jobs through the "Job Composer" in the jobs dropdown. | |||
[[File:OOD JOBS COMPOSER NEW.png|Screenshot showing the ability to create new jobs within the ood job composer]] | |||
=== RStudio === | If we create a new job from a template, you're given a list of templates to use: | ||
[[File:OOD JOBS COMPOSER NEW FROM TEMPLATE.png|Screenshot of some example templates for jobs within openondemand]] | |||
If we choose the default template, you can run it, or edit the job script to make it do what you would like | |||
[[File:OOD JOBS COMPOSER SUBMIT.png|Screenshot showing the ability to submit or edit jobs within the Job composer in OpenOnDemand]] | |||
=== Interactive Applications === | |||
We have a number of interactive applications being made available through OpenOnDemand | |||
* Beocat Desktop | |||
* [https://www.comsol.com/ COMSOL] | |||
* [https://www.gnu.org/software/octave/ Octave] | |||
* [https://www.ks.uiuc.edu/Research/vmd/ VMD] | |||
* [https://www.wolfram.com/mathematica/ Mathematica] Please note, this is from a site license limited to KSU students, faculty and staff. | |||
* [https://afni.nimh.nih.gov/ AFNI] | |||
* [https://coder.com/ CodeServer] is a cloud native version of VS Code that runs on the compute nodes. Useful, since VS Code's remote connections cannot be used with Beocat. Other names for VS Code may be VSCode or Visual Studio Code. | |||
* [https://jupyter.org/ Jupyter] | |||
* [https://www.rstudio.com/products/rstudio-server/ RStudio] | |||
==== RStudio ==== | |||
RStudio is one of the interactive applications that we've enabled for use within OpenOnDemand | RStudio is one of the interactive applications that we've enabled for use within OpenOnDemand | ||
| Line 39: | Line 74: | ||
[[File:Ood RStudio.png|Screenshot showing RStudio through OpenOnDemand]] | [[File:Ood RStudio.png|Screenshot showing RStudio through OpenOnDemand]] | ||
=== Jupyter === | ==== Jupyter ==== | ||
Like RStudio above, click on Interactive Apps and then go to Jupyter. From there, you'll have a form that allows you to specify requirements for your Jupyter run. | |||
[[File:OOD JUPYTER LAUNCH.png|Screenshot of options to launch Jupyter]] | |||
Once the job is launched it will take you to a page where you can connect to your running Jupyter service | |||
[[File:Ood INTERACTIVE APPS JUPYTER.png|Screenshot of connection option for jupyter]] | |||
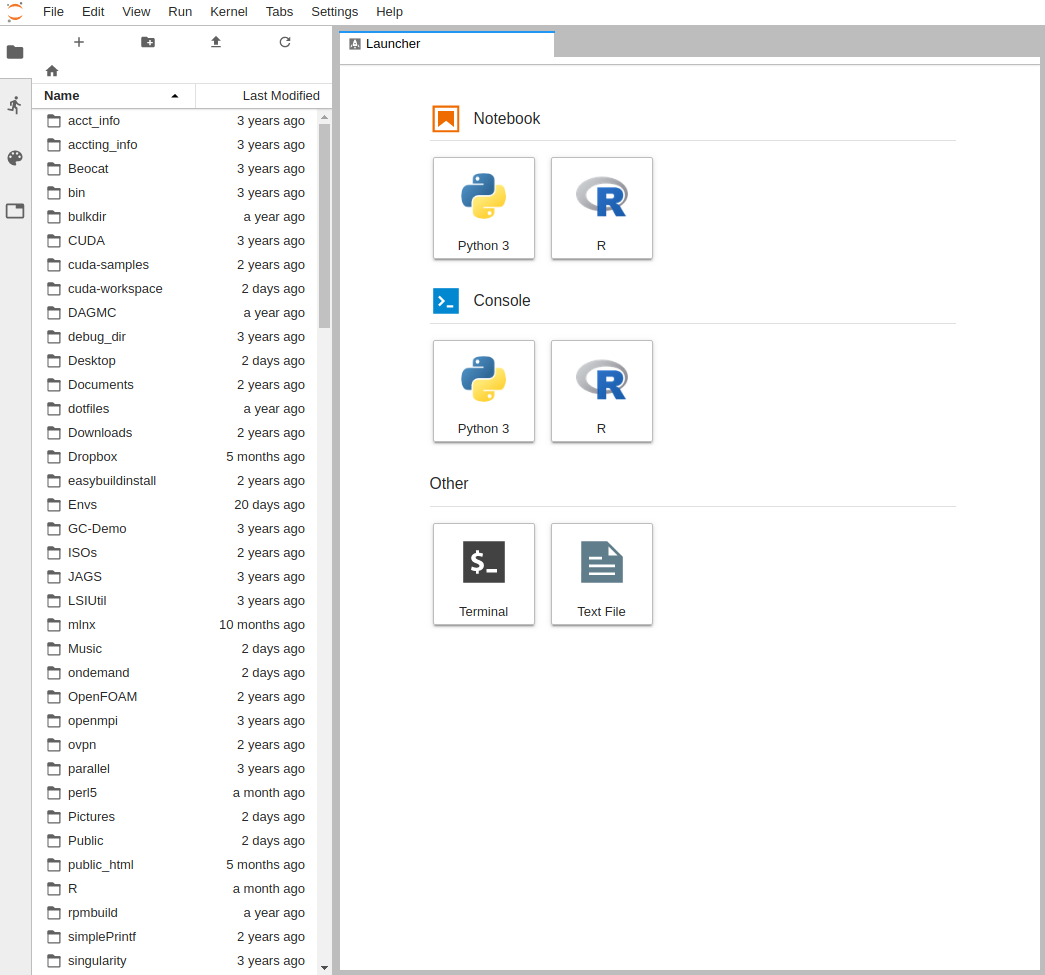
It will then take you to the interface you chose, below is the JupyterLab interface: | |||
[[File:OOD JUPYTER LAB.png|Screenshot of JupyterLab through OpenOnDemand]] | |||
Jupyter Kernels currently supported: | |||
* Python 2 | |||
* Python 3 | |||
* R | |||
* Octave | |||
* Sage | |||
Julia support will come, but will need each user to set it up individually. There is currently a large bug centered around Julia, CentOS/RHEL, and our shared filesystem (Ceph). | |||
===== Installing python libraries ===== | |||
You may need to install extra python libraries to use with your Jupyter Python kernels. For instance, this is how you'd install tobler from within a Jupyter Notebook: | |||
<syntaxhighlight lang=python> | |||
!pip install --user tobler | |||
# Sometimes Jupyter notebook needs to then be told how to find the libraries you've installed in that manner. | |||
# Your username should be put in place of the <PUT_YOUR_USERNAME_HERE> text. | |||
# this will need to change if you are not using a 3.7 kernel | |||
import sys | |||
sys.path.append("/homes/<PUT_YOUR_USERNAME_HERE>/.local/lib/python3.7/site-packages") | |||
</syntaxhighlight> | |||
===== Creating python virtual environments ===== | |||
You can create separate python virtual libraries for each project to keep the libraries separate for each. | |||
Each virtual environment can be created using the commands below in a terminal on Beocat, or you can | |||
precede each with an exclamation point and issue them from within a Jupyter notebook. | |||
<syntaxhighlight lang=bash> | |||
# First we'll activate the ability to use the ondemand modules: | |||
module use /opt/beocat/ondemand_modules | |||
# Then we can list the jupyter_python modules so we can use them to create the virtual environment: | |||
module avail jupyter_python | |||
# Load the version you would like (ideally you use a jupyter_python module for this, otherwise the virtual environment itself will have to have a decent amount of jupyter libraries installed into it) | |||
module load jupyter_python/3.10.4-TensorFlow-2.11.0 | |||
# If you'd like to see what libraries this actually loaded, you can check it with the following: | |||
module list | |||
# We'll create a virtual environment (activate it and install any libraries you need. | |||
python -m venv --system-site-packages /homes/mozes/virtualenvs/testing_ondemand_jupyter | |||
. /homes/mozes/virtualenvs/testing_ondemand_jupyter/bin/activate | |||
pip install # insert needed libraries here | |||
# here we create a directory to hold the configuration files for telling our Jupyter environment about your virtual environment | |||
mkdir -p ~/ondemand/jupyter_kernel_configs | |||
# Now we need to create a configuration file to instruct Jupyter to find this virtual environment | |||
nano ~/ondemand/jupyter_kernel_configs/my_environment_name.sh | |||
# in that file should be lines like the following: | |||
NAME="testing_ondemand_virtualenv" | |||
VIRTUAL_ENV="/homes/mozes/virtualenvs/testing_ondemand_jupyter" | |||
MODULES="jupyter_python/3.10.4-TensorFlow-2.11.0" | |||
# of course, you should provide your own name and path to the virtual environment. Please don't put spaces in the name. | |||
</syntaxhighlight> | |||
Once you start a new jupyter session it should list the new kernel option that uses your virtual environment. | |||
From the '''Jupyter''' page with the '''Files''' menu chosen you should find your kernel under the '''New''' | |||
menu on the top right. | |||
===== Installing libraries with conda ===== | |||
Conda environments should automatically show up if ''conda'' is in the PATH. For them to function properly, they will need at least the ipykernel library installed into them. | |||
conda install -c conda-forge ipykernel | |||
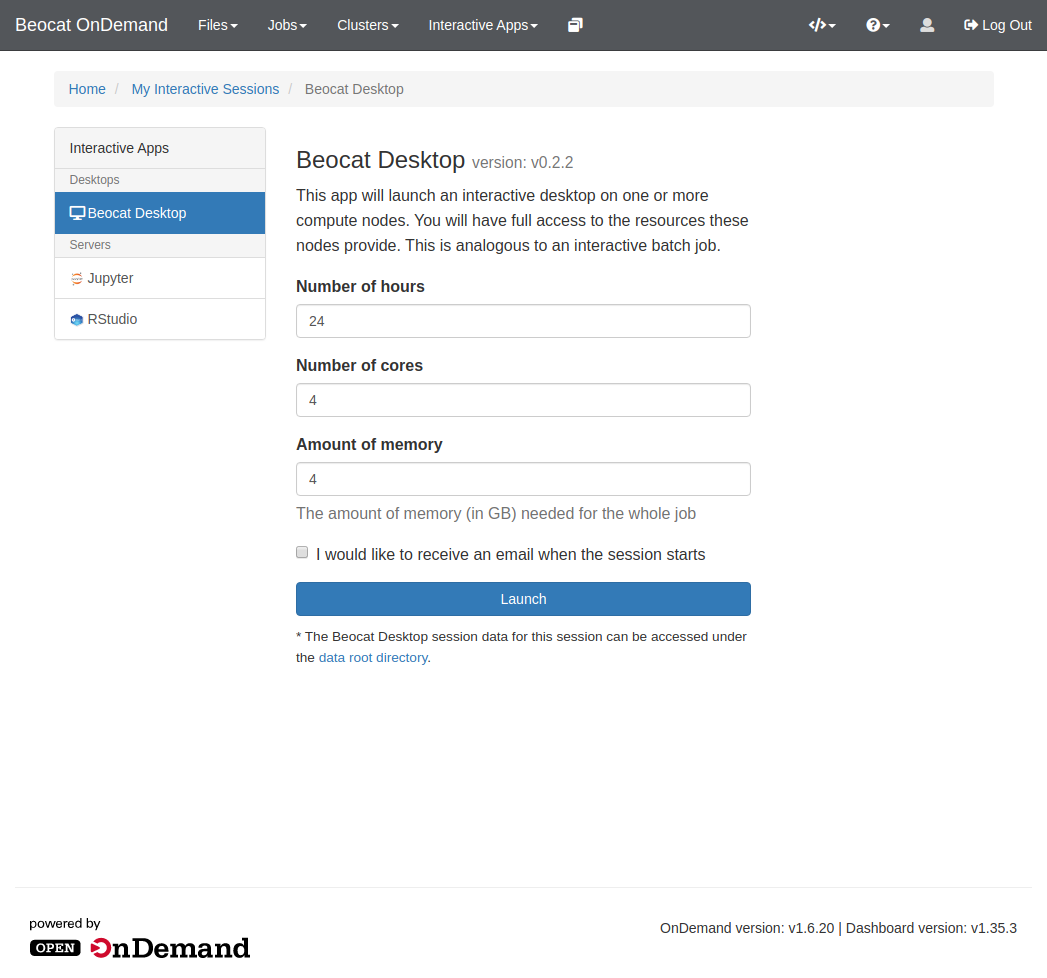
==== Beocat Desktop ==== | |||
Sometimes, you just need a Desktop somewhere to run your graphical applications. This can be done through the Beocat Desktop option in the Interactive Apps dropdown on the dashboard. | |||
[[File:OOD DESKTOP LAUNCH.png|Screenshot of the options to launch a graphical desktop through OpenOnDemand]] | |||
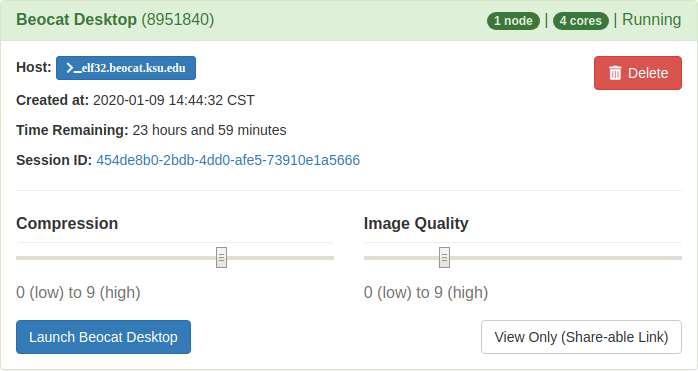
Once launched, you'll be able to connect to the desktop through vnc in the openondemand in the "My Interactive Sessions" tab. | |||
[[File:OOD INTERACTIVE APPS DESKTOP.png|Screenshot of VNC options for Desktop in OpenOnDemand]] | |||
Once you've launched the Beocat Desktop, you can interact with it like a normal desktop through the browser. | |||
[[File:OOD DESKTOP VNC.png|Screenshot of VNC Beocat Desktop]] | |||
=== Shell Access === | === Shell Access === | ||
Latest revision as of 13:27, 3 April 2025
OpenOnDemand
OpenOnDemand is a platform for running computational tasks on a cluster from a web browser. If those tasks are interactive, it provides the ability to interact with them once the task has started its execution. OpenOnDemand has an "App" based plugin system for adding new types of computational tasks and interactivity.
One of the greatest benefits of this system is remote access to large machines for computational tasks, without the need for utilizing a command-line interface that is difficult to learn.
Our installation is available at https://ondemand.beocat.ksu.edu
File Management
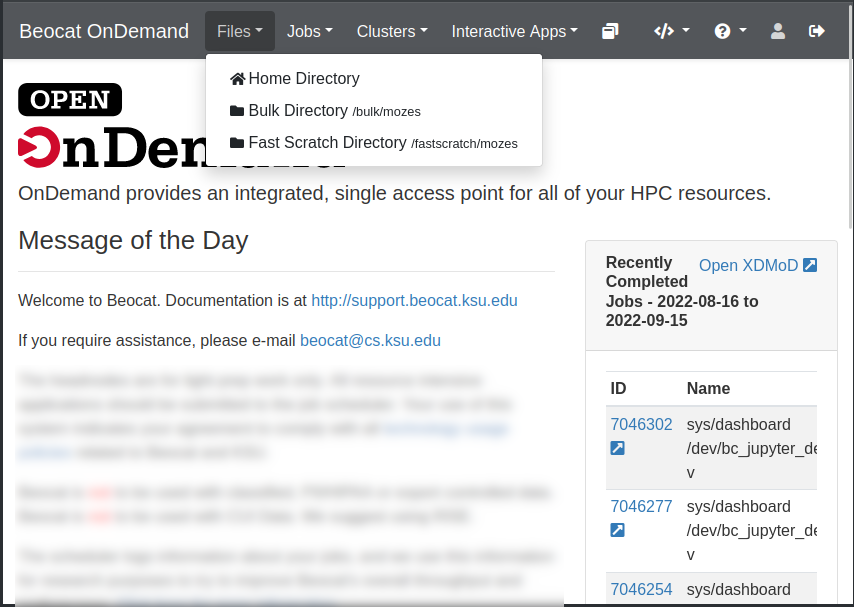
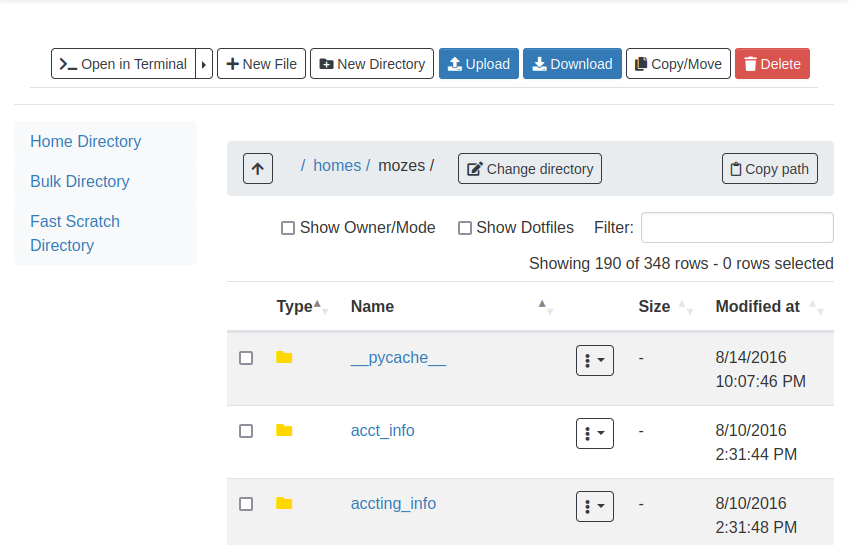
File management can be accessed through the Files dropdown in the dashboard.
Once you click on Home Directory, you can manage your files, upload/download/rename/edit and view.
If you're looking for a way to get your files into and out of OneDrive, Google Drive, other cloud providers you may be interested in taking a look at our documentation for Onedrive Data Transfer
Job Management
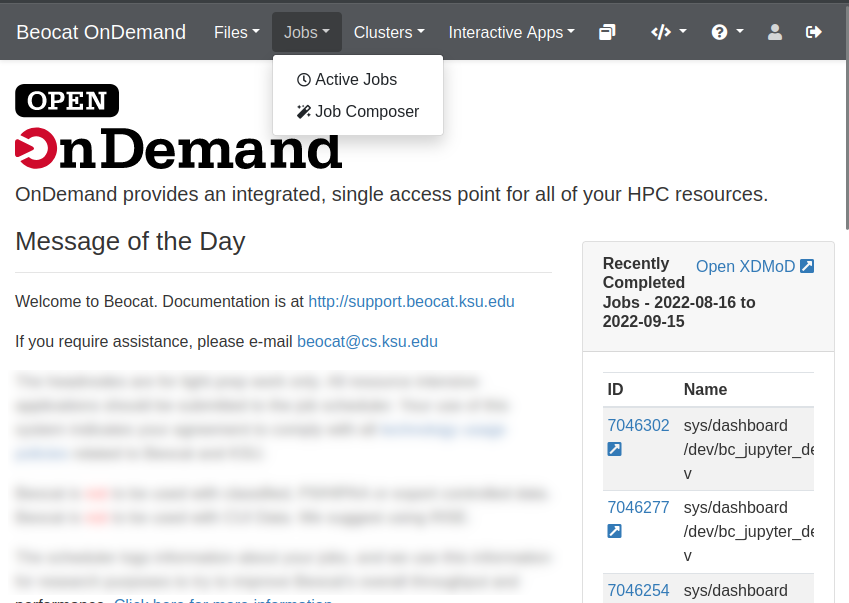
A cluster isn't much of a cluster if it can't run jobs for you to lookup later. OpenOnDemand has a robust job management application builtin.
It is accessible from the Jobs dropdown in the dashboard.
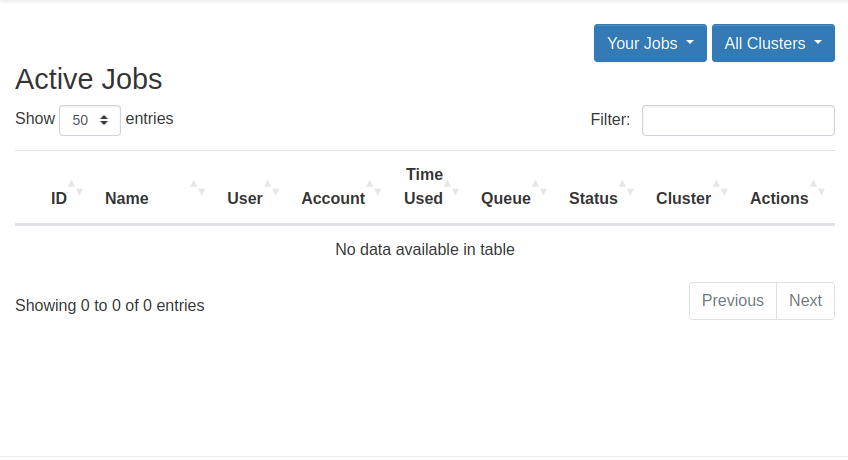
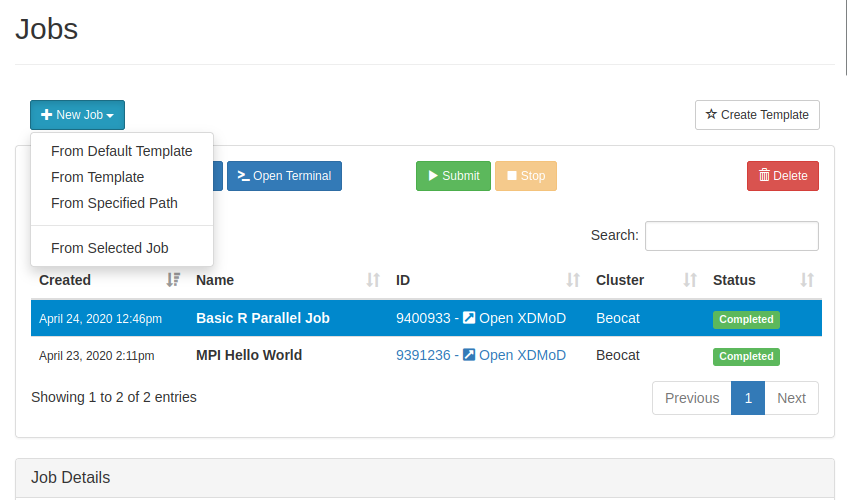
View Active Jobs
You can view your active jobs and get more information about them from the Active Jobs option in the Jobs dropdown
Compose Jobs
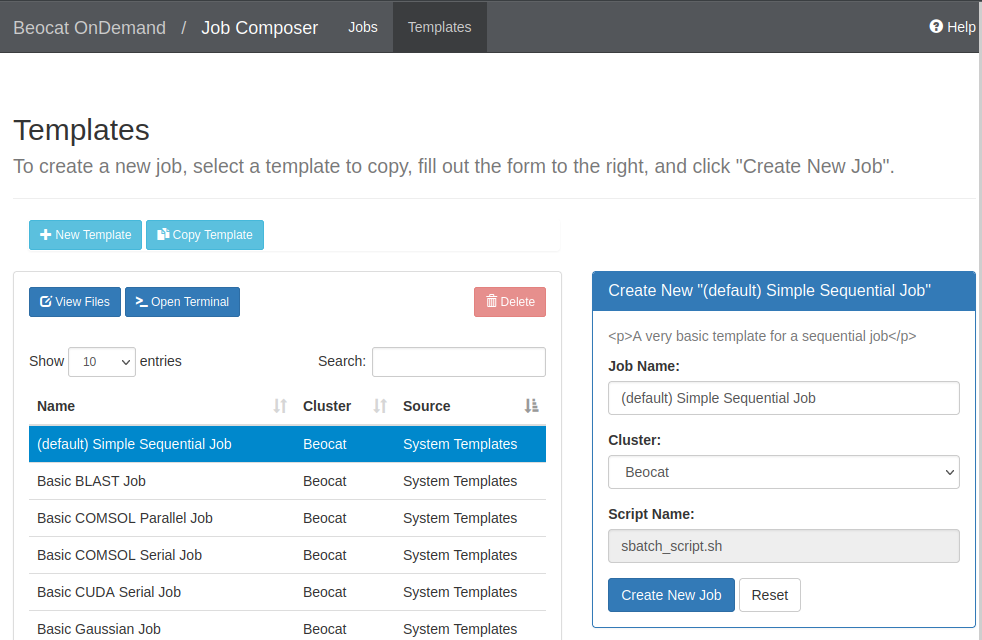
You can create new jobs through the "Job Composer" in the jobs dropdown.
If we create a new job from a template, you're given a list of templates to use:
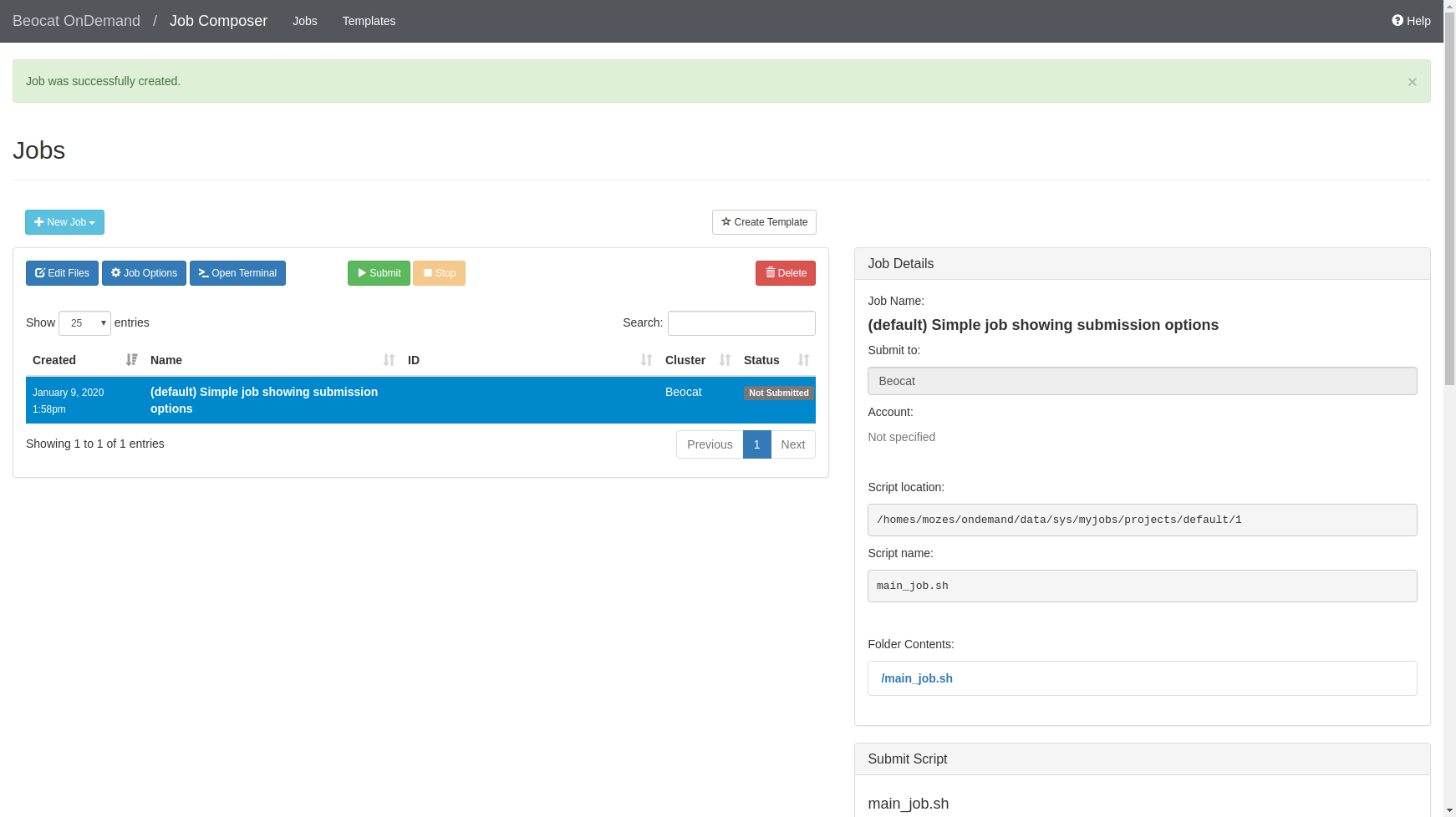
If we choose the default template, you can run it, or edit the job script to make it do what you would like
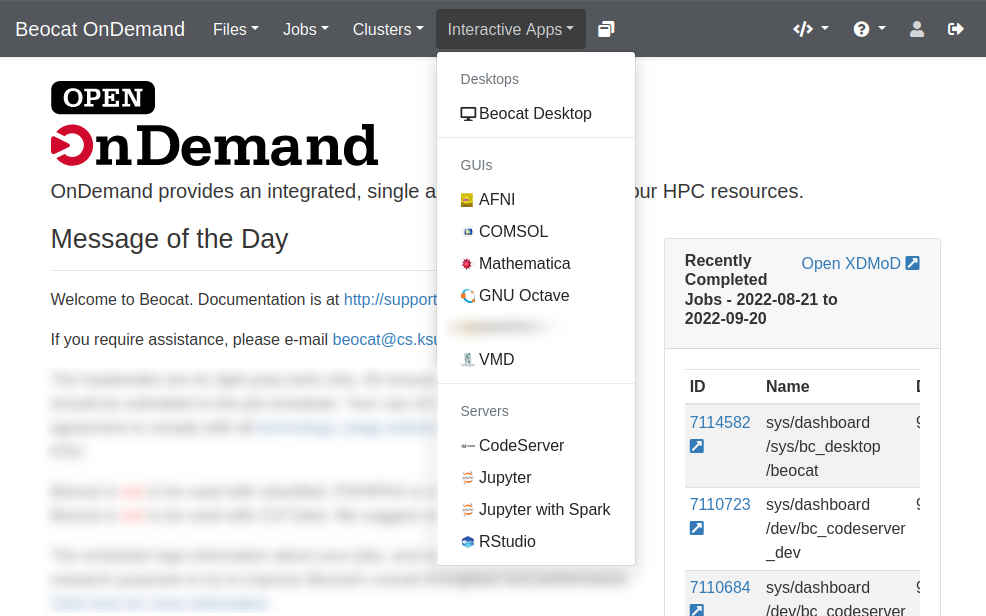
Interactive Applications
We have a number of interactive applications being made available through OpenOnDemand
- Beocat Desktop
- COMSOL
- Octave
- VMD
- Mathematica Please note, this is from a site license limited to KSU students, faculty and staff.
- AFNI
- CodeServer is a cloud native version of VS Code that runs on the compute nodes. Useful, since VS Code's remote connections cannot be used with Beocat. Other names for VS Code may be VSCode or Visual Studio Code.
- Jupyter
- RStudio
RStudio
RStudio is one of the interactive applications that we've enabled for use within OpenOnDemand
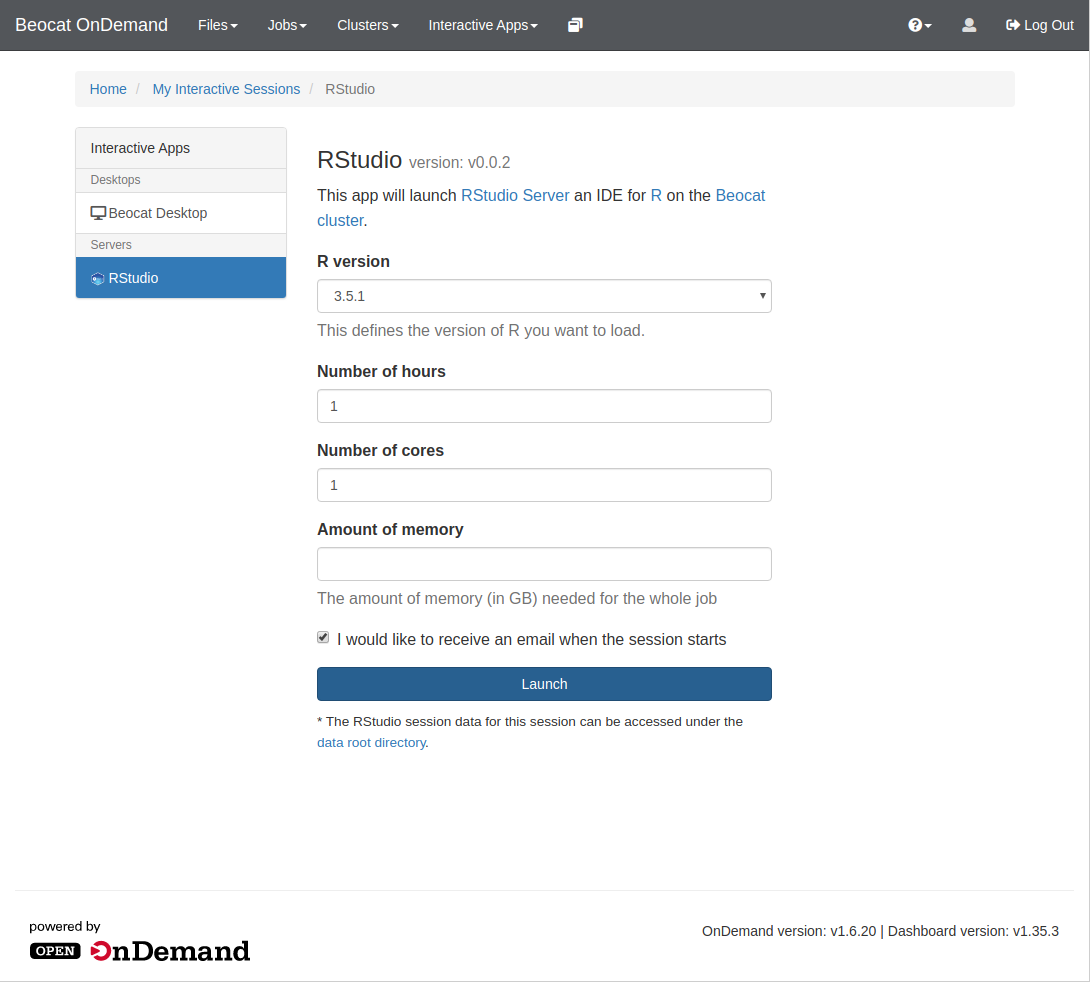
You launch interactive apps through the "Interactive Apps" dropdown.
Once you click on RStudio, you'll be brought to a page allowing you to specify requirements for your RStudio run, e.g. memory, cores, and runtime.
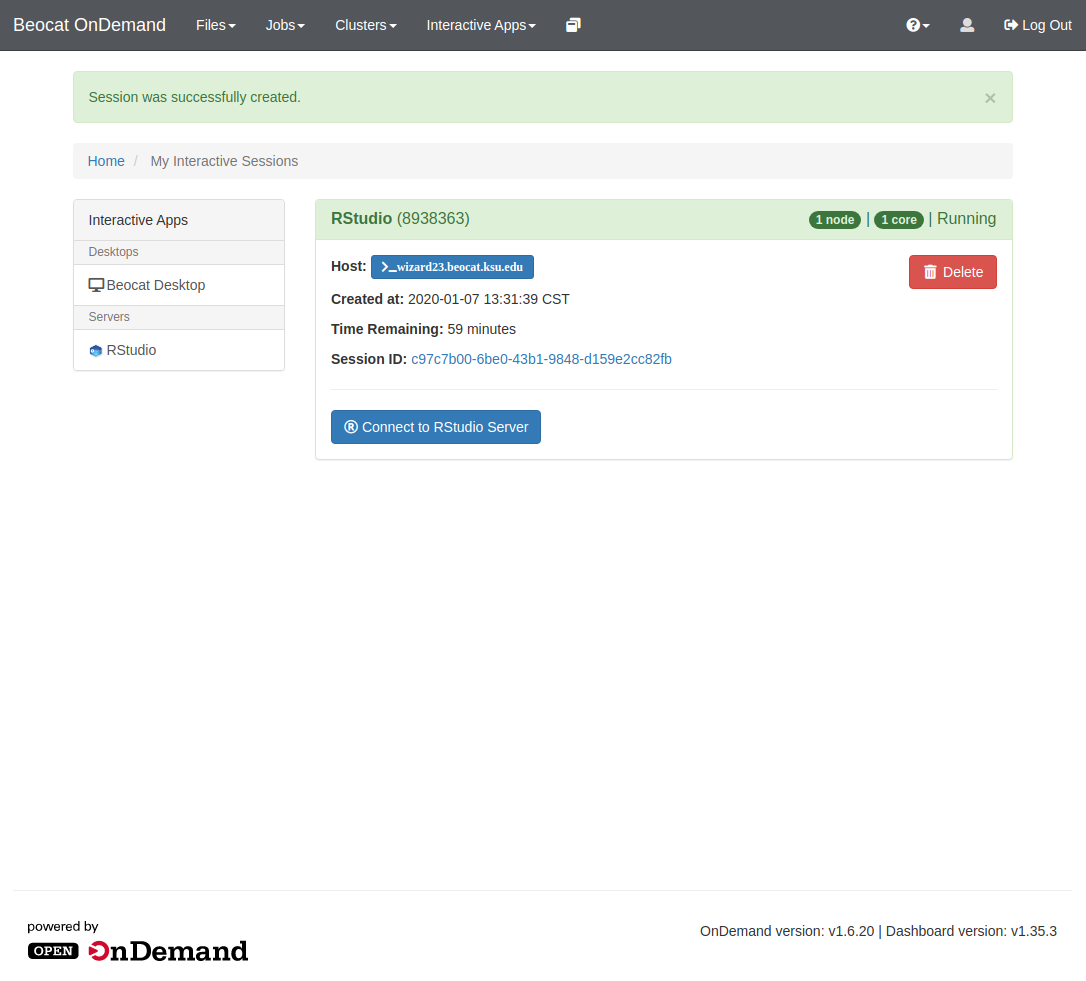
Once the job is submitted, the scheduler will take it and run it when space is available. Once the job is running, "My Interactive sessions" page will look like this:
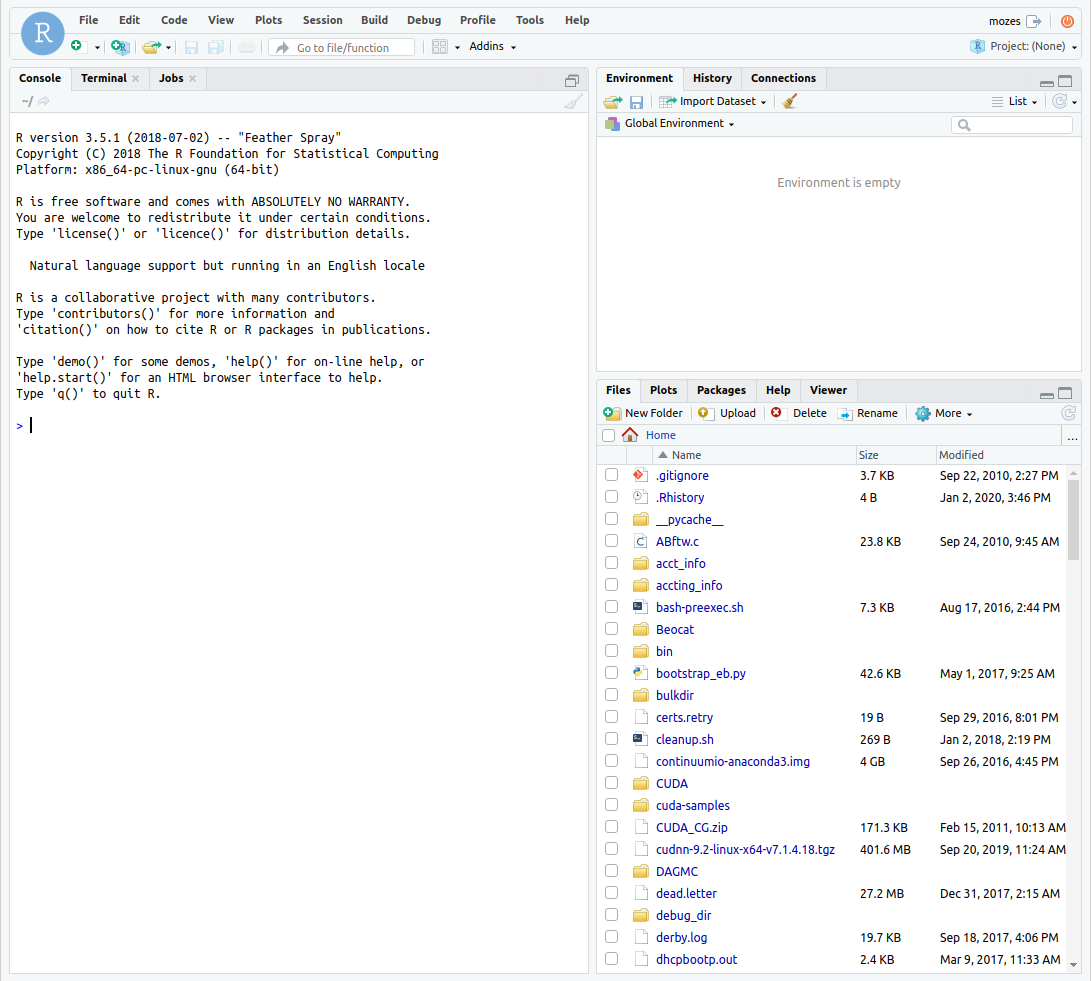
From there, you can connect to RStudio and it will bring you to a familiar interface.
Jupyter
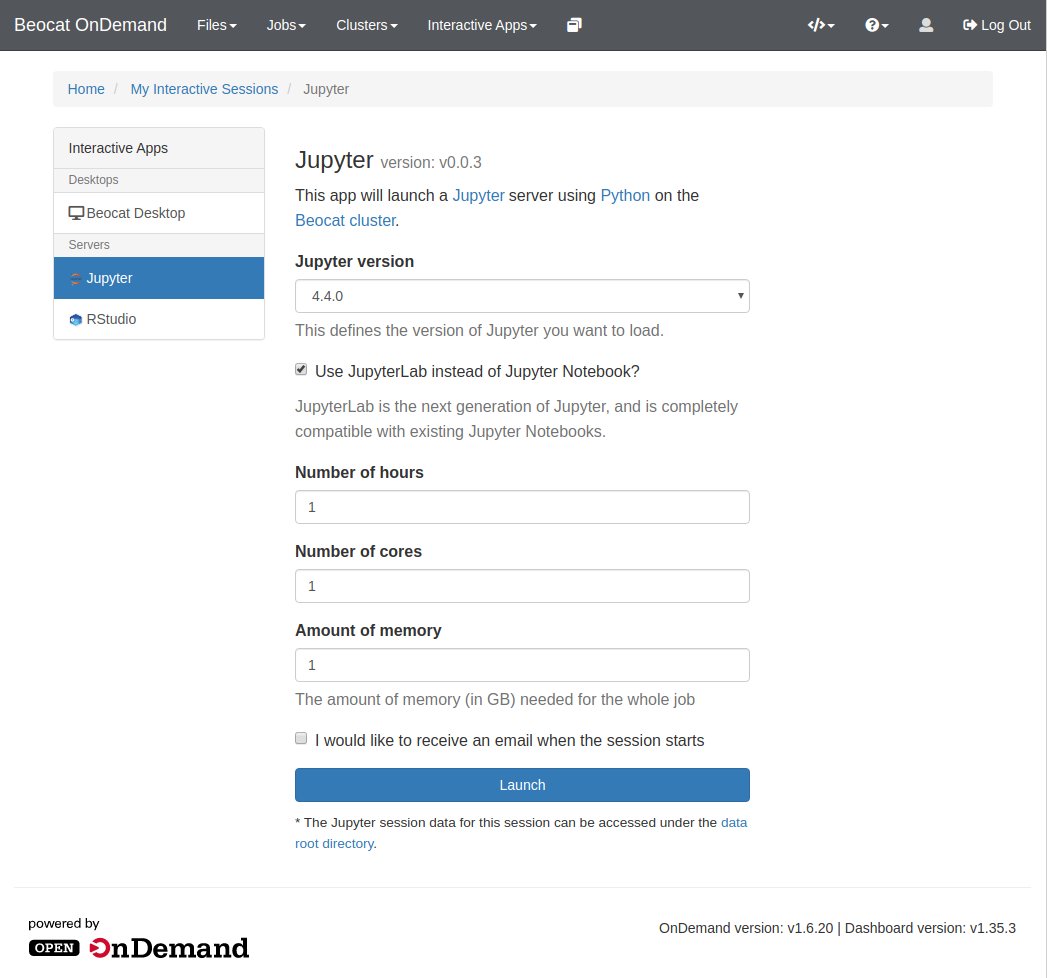
Like RStudio above, click on Interactive Apps and then go to Jupyter. From there, you'll have a form that allows you to specify requirements for your Jupyter run.
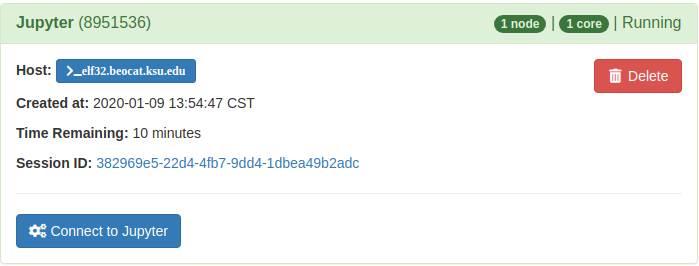
Once the job is launched it will take you to a page where you can connect to your running Jupyter service
It will then take you to the interface you chose, below is the JupyterLab interface:
Jupyter Kernels currently supported:
- Python 2
- Python 3
- R
- Octave
- Sage
Julia support will come, but will need each user to set it up individually. There is currently a large bug centered around Julia, CentOS/RHEL, and our shared filesystem (Ceph).
Installing python libraries
You may need to install extra python libraries to use with your Jupyter Python kernels. For instance, this is how you'd install tobler from within a Jupyter Notebook:
!pip install --user tobler
# Sometimes Jupyter notebook needs to then be told how to find the libraries you've installed in that manner.
# Your username should be put in place of the <PUT_YOUR_USERNAME_HERE> text.
# this will need to change if you are not using a 3.7 kernel
import sys
sys.path.append("/homes/<PUT_YOUR_USERNAME_HERE>/.local/lib/python3.7/site-packages")
Creating python virtual environments
You can create separate python virtual libraries for each project to keep the libraries separate for each. Each virtual environment can be created using the commands below in a terminal on Beocat, or you can precede each with an exclamation point and issue them from within a Jupyter notebook.
# First we'll activate the ability to use the ondemand modules:
module use /opt/beocat/ondemand_modules
# Then we can list the jupyter_python modules so we can use them to create the virtual environment:
module avail jupyter_python
# Load the version you would like (ideally you use a jupyter_python module for this, otherwise the virtual environment itself will have to have a decent amount of jupyter libraries installed into it)
module load jupyter_python/3.10.4-TensorFlow-2.11.0
# If you'd like to see what libraries this actually loaded, you can check it with the following:
module list
# We'll create a virtual environment (activate it and install any libraries you need.
python -m venv --system-site-packages /homes/mozes/virtualenvs/testing_ondemand_jupyter
. /homes/mozes/virtualenvs/testing_ondemand_jupyter/bin/activate
pip install # insert needed libraries here
# here we create a directory to hold the configuration files for telling our Jupyter environment about your virtual environment
mkdir -p ~/ondemand/jupyter_kernel_configs
# Now we need to create a configuration file to instruct Jupyter to find this virtual environment
nano ~/ondemand/jupyter_kernel_configs/my_environment_name.sh
# in that file should be lines like the following:
NAME="testing_ondemand_virtualenv"
VIRTUAL_ENV="/homes/mozes/virtualenvs/testing_ondemand_jupyter"
MODULES="jupyter_python/3.10.4-TensorFlow-2.11.0"
# of course, you should provide your own name and path to the virtual environment. Please don't put spaces in the name.
Once you start a new jupyter session it should list the new kernel option that uses your virtual environment. From the Jupyter page with the Files menu chosen you should find your kernel under the New menu on the top right.
Installing libraries with conda
Conda environments should automatically show up if conda is in the PATH. For them to function properly, they will need at least the ipykernel library installed into them.
conda install -c conda-forge ipykernel
Beocat Desktop
Sometimes, you just need a Desktop somewhere to run your graphical applications. This can be done through the Beocat Desktop option in the Interactive Apps dropdown on the dashboard.
Once launched, you'll be able to connect to the desktop through vnc in the openondemand in the "My Interactive Sessions" tab.
Once you've launched the Beocat Desktop, you can interact with it like a normal desktop through the browser.
Shell Access
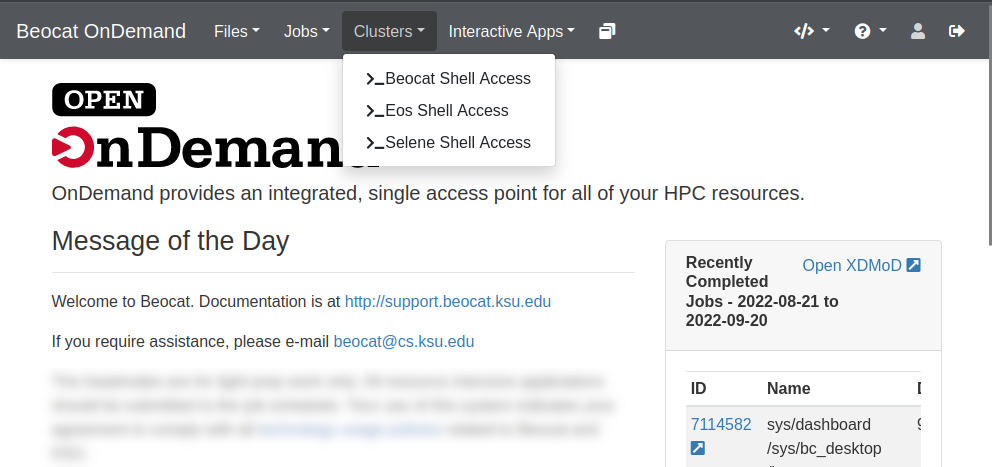
Somethings, no matter how hard we try, are easier to do via the command line. OpenOnDemand also gives you a way to handle those cases via the Clusters dropdown
You can choose an individual headnode, if need-be, or you can choose "Beocat Shell Access" to be given one of the headnodes at random. Once chosen, you should be able to have a familiar command-line experience